Note
Go to the end to download the full example code.
PCA via SVD and via AffinityMatcher#
We show how to compute a PCA embedding using the closed form and using the AffinityMatcher class. Both approaches lead to the same solution.
# Author: Hugues Van Assel <vanasselhugues@gmail.com>
#
# License: BSD 3-Clause License
import matplotlib.pyplot as plt
from sklearn.datasets import load_digits
import torch
from torchdr import AffinityMatcher, PCA
from torchdr.affinity import Affinity
class ScalarProductAffinity(Affinity):
"""Compute the scalar product affinity matrix X @ X.T."""
def __init__(self, device="auto", backend=None, verbose=False):
super().__init__(
metric="angular",
device=device,
backend=backend,
verbose=verbose,
zero_diag=False,
)
def _compute_affinity(self, X):
return X @ X.T
Load toy images#
First, let’s load 5 classes of the digits dataset from sklearn.
digits = load_digits(n_class=5)
X = digits.data
X = X - X.mean(0)
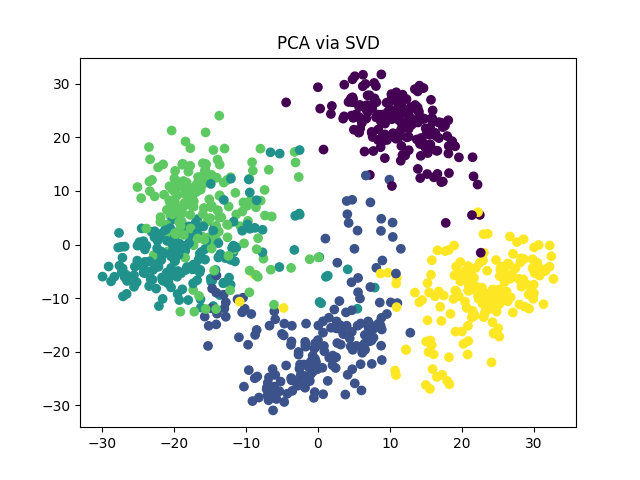
PCA via SVD#
Let us perform PCA using the closed form solution given by the
Singular Value Decomposition (SVD).
In Torchdr, it is available at torchdr.PCA.
PCA via AffinityMatcher#
Now, let us perform PCA using the AffinityMatcher class
torchdr.AffinityMatcher
as well as the scalar product affinity
torchdr.ScalarProductAffinity
for both input data and embeddings,
and the square loss as global objective.
model = AffinityMatcher(
n_components=2,
affinity_in=ScalarProductAffinity(),
affinity_out=ScalarProductAffinity(),
loss_fn="square_loss",
init="normal",
lr=1e1,
max_iter=50,
backend=None,
)
Z_am = model.fit_transform(X)
plt.figure()
plt.scatter(Z_am[:, 0], Z_am[:, 1], c=digits.target)
plt.title("PCA via AffinityMatcher")
plt.show()
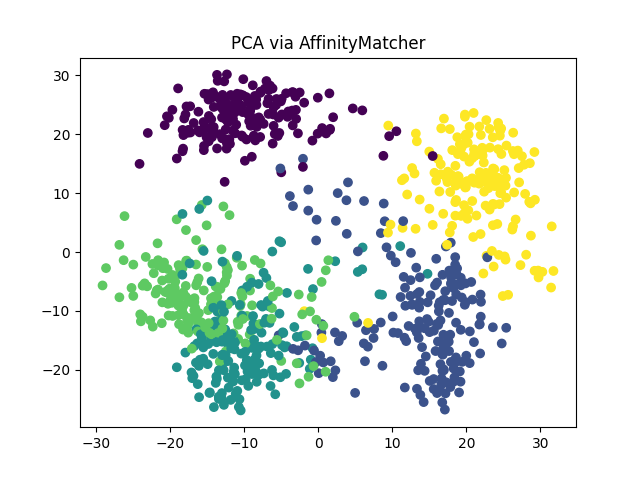
We can see that we obtain the same PCA embedding (up to a rotation) using both methods.
Total running time of the script: (0 minutes 3.932 seconds)
