Note
Go to the end to download the full example code.
Neighbor Embedding on genomics & equivalent affinity matcher formulation#
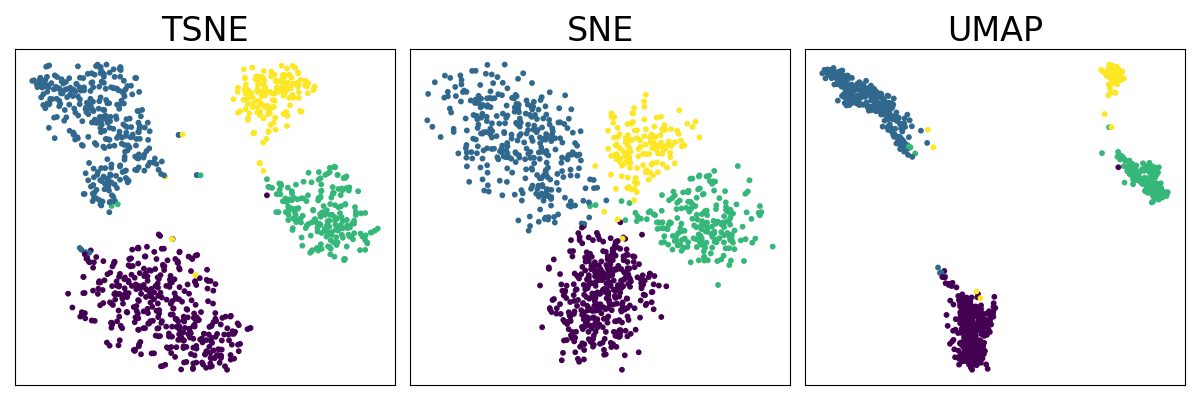
We illustrate the basic usage of TorchDR with different neighbor embedding methods on the SNARE-seq gene expression dataset with given cell type labels.
# Author: Titouan Vayer <titouan.vayer@inria.fr>
# Hugues Van Assel <vanasselhugues@gmail.com>
#
# License: BSD 3-Clause License
import urllib.request
import matplotlib.pyplot as plt
import numpy as np
from torchdr import (
SNE,
TSNE,
UMAP,
AffinityMatcher,
EntropicAffinity,
NormalizedGaussianAffinity,
)
Load the SNARE-seq dataset (gene expression) with cell type labels#
def load_numpy_from_url(url, delimiter="\t"):
response = urllib.request.urlopen(url)
data = response.read().decode("utf-8")
data = data.split("\n")
data = [row.split(delimiter) for row in data if row]
numpy_array = np.array(data, dtype=float)
return numpy_array
url_x = "https://rsinghlab.github.io/SCOT/data/snare_rna.txt"
X = load_numpy_from_url(url_x)
url_y = "https://rsinghlab.github.io/SCOT/data/SNAREseq_types.txt"
Y = load_numpy_from_url(url_y)
Run neighbor embedding methods#
params = {
"optimizer": "Adam",
"optimizer_kwargs": None,
"max_iter": 100,
"lr": 1e0,
"backend": None,
}
sne = SNE(early_exaggeration_coeff=1, **params)
umap = UMAP(**params)
tsne = TSNE(early_exaggeration_coeff=1, **params)
all_methods = {
"TSNE": tsne,
"SNE": sne,
"UMAP": umap,
}
for method_name, method in all_methods.items():
print(f"--- Computing {method_name} ---")
method.fit(X)
--- Computing TSNE ---
--- Computing SNE ---
--- Computing UMAP ---
Plot the different embeddings#
fig = plt.figure(figsize=(12, 4))
for i, (method_name, method) in enumerate(all_methods.items()):
ax = fig.add_subplot(1, 3, i + 1)
emb = method.embedding_.detach().numpy() # get the embedding
ax.scatter(emb[:, 0], emb[:, 1], c=Y, s=10)
ax.set_title(f"{method_name}", fontsize=24)
ax.set_xticks([])
ax.set_yticks([])
plt.tight_layout()
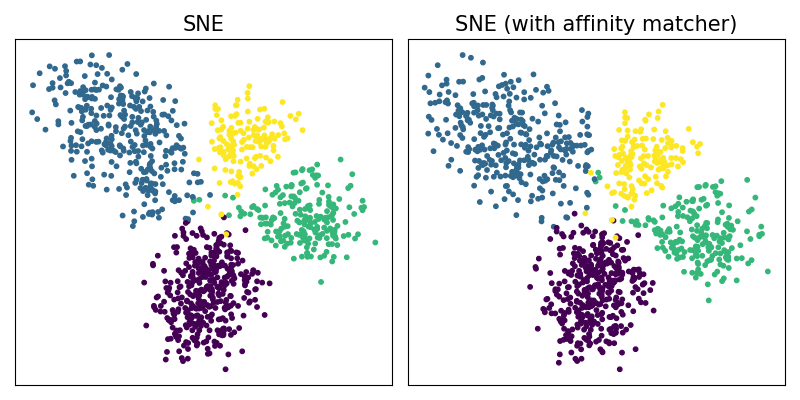
Using AffinityMatcher#
We can reproduce the same embeddings using the
class torchdr.AffinityMatcher. The latter takes as input
two affinities and minimize a certain matching loss between them.
To reproduce the SNE algorithm
we can match, via the cross entropy loss,
a torchdr.EntropicAffinity with a
torchdr.NormalizedGaussianAffinity.
sne_affinity_matcher = AffinityMatcher(
n_components=2,
# SNE matches an EntropicAffinity
affinity_in=EntropicAffinity(sparsity=False),
# with a Gaussian kernel normalized by row
affinity_out=NormalizedGaussianAffinity(normalization_dim=1),
loss_fn="cross_entropy_loss", # and the cross_entropy loss
**params,
)
sne_affinity_matcher.fit(X)
fig = plt.figure(figsize=(8, 4))
two_sne_dict = {"SNE": sne, "SNE (with affinity matcher)": sne_affinity_matcher}
for i, (method_name, method) in enumerate(two_sne_dict.items()):
ax = fig.add_subplot(1, 2, i + 1)
emb = method.embedding_.detach().numpy() # get the embedding
ax.scatter(emb[:, 0], emb[:, 1], c=Y, s=10)
ax.set_title(f"{method_name}", fontsize=15)
ax.set_xticks([])
ax.set_yticks([])
plt.tight_layout()
On the efficiency of using the torchdr API rather than AffinityMatcher directly#
Note
Calling torchdr.SNE enables to leverage sparsity and therefore
significantly reduces the computational cost of the algorithm compared to
using torchdr.AffinityMatcher with the corresponding affinities.
In TorchDR, it is therefore recommended to use the specific class associated
with the desired algorithm when available.
Total running time of the script: (1 minutes 39.774 seconds)
